
|
Introduction 2-Aminophenol is an organic compound with molecular formula C6H4(OH)NH2. It is an amphoteric molecule and a reducing agent useful for the synthesis of dyes and heterocyclic compounds [1]. 2-Aminophenol can be prepared via the reduction of 2-nitrophenol, and it has a rather high melting point due to internal hydrogen bonding (m.p. 174 °C) [2]. A computational study on the enhanced stabilization of aminophenol derivatives by internal hydrogen bonding has been reported [3]. Jose et al. have conducted the density functional theory (DFT) study on the thermodynamic properties of aminophenols [4]. Experimental and theoretical investigation of first hyperpolarizability in aminophenols have been performed and reported by Franzen et al. [5]. In the present study, the vibrational and NMR spectra of the title compound have been calculated and compared with experimental results [10]. Computational Details Calculations of the title compound were carried out with Gaussian 03™ software program [6]. The structure of 2-aminophenol has been optimized at two levels of theory using two basis sets. They are RHF/6-31 G (d), RHF/cc-pVDZ, B3LYP/6-31 G(d) and B3LYP/cc-pVDZ [7, 8]. Molecular geometries were fully optimized by Berny’s optimization algorithm using redundant internal coordinates. The structural parameters of 2-aminophenol determined by these calculations are listed in Tables 1 and 2 in accordance with the atom numbering scheme in Figure 1. Tables 1 and 2 compare the calculated bond lengths and angles for 2-aminophenol with those of experimentally available data [10]. Using the optimized geometry, the vibrational frequencies were calculated using the same level of theory and the same basis sets. Harmonic vibrational wavenumbers were calculated using analytic second derivatives to confirm convergence to minima on the potential surface.
At the optimized structure (Figure 1) of the examined species, no imaginary wave number modes were obtained, proving that a true minimum on the potential surface was found. The proton NMR and C13 NMR of the title compound have been calculated using the gauge independent atomic orbitals (GIAO) method. Results & Discussion Bond Length All C-C bond lengths in 2-aminophenol have the same value in both HF and DFT methods. The C-H bond length exhibits slight variations when basis set changes from 6-31G (d) to cc-pVDZ in the Hartee-Fock (HF) method. The same bond length has unaltered values in the DFT method. N-H bond length has the same values in DFT method for both basis sets, whereas in HF method bond length increases by 0.01Å as the basis set changes from 6-31G(d) to cc-pVDZ. C=O bond length has same values in the HF and DFT methods. The C=O bond length increases by 0.01Å as the theory changes from RHF to DFT. O-H bond length has same values in HF and DFT method as basis set varies from 6-31G(d) to cc-pVDZ. O-H bond length increases by 0.03Å as theory changes from RHF to DFT. The bond lengths of 2-amino phenol are given in Table 1. Bond Angles In 2-aminophenol the bond angle values obtained by RHF and B3LYP calculations exhibit a slight difference in 6-31G(d) and cc-pVDZ basis sets. But in some cases, like C1-C2-N11, C1-O7-H8, C6-C1-O7, C4-C5-H14, there is much difference in bond angles of RHF and B3LYP calculations. The bond angles of 2-amino phenol are given in Table 2. Vibrational Frequencies The vibrational frequencies were calculated computationally using two basis sets 6-31G(d) and cc-pVDZ and two theories RHF and B3LYP. The DFT hybrid B3LYP tends to overestimate the fundamental modes, therefore scaling factors have to be used for obtaining a considerably better agreement with experimental data. Therefore, a scaling factor of 0.9613 and 0.8929 was uniformly applied to the wavenumbers calculated using DFT and HF, as suggested in Gaussian 03™ [9]. The vibrational frequency obtained from the calculation using RHF method was multiplied by a factor 0.8929 and by 0.9613 in DFT method for scaling. The frequencies are then being compared with the experimental spectrum obtained from the SDBS website [10]. The computational and experimental frequencies of 2-aminophenol with their assignments are given in Table 3 [11, 12].
H1 NMR The proton NMR of the title compound has been calculated using the gauge independent atomic orbitals (GIAO) method [13, 14]. From the isotropic values of the molecule and that of TMS we obtain the δ values of the nmr spectrum. The isotropic value of the protons of TMS is 31.6198 in the RHF calculation and 31.7825 in B3LYP calculation. The proton-nmr spectrum of 2-aminophenol is taken from the website SDBS [10]. The δ values of 2-aminophenol are given in Table 4.
The proton-nmr spectrum of 2-aminophenol exhibits six peaks. This indicates six types of protons. The computationally calculated values of protons 8, 9, 12, 10, 15, 13 and 14 agree with the experimental values, but the computationally calculated values of protons 9 and 12 are less than the experimental values in both HF and DFT methods. The chemical shift values calculated by GIAO method show slight difference from the experimental values, but maintain a general trend of chemical shift. C13 NMR The isotropic value of the C13 of TMS is 195.1196 in the RHF theory and 188.7879 in the theory B3LYP. The C13 nmr spectrum of the molecule is taken from the website SDBS [10]. From the isotropic values of the carbon of 2-aminophenol and the TMS we obtain the δ values of their C13 nmr spectrum. The δ values of 2-aminophenol are given in Table 5. The C13 spectrum of 2-aminophenol reveals 6 peaks, which indicates 6 different types of carbon atoms. From the computational calculations also we have 6 values which agree with the experimental values.
HOMO-LUMO energy gaps The relative energy of the molecular orbitals have been calculated and a graphical representation of the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) of 2-aminophenol are given in Figures 2(a) and (b). 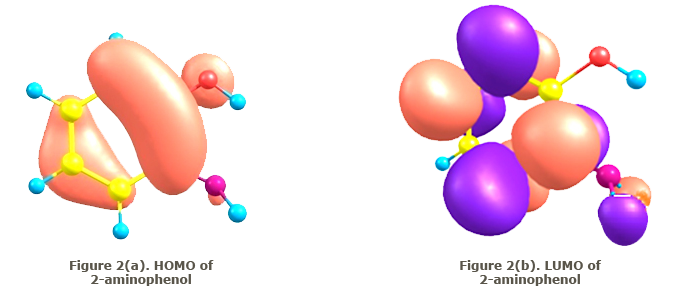 Conclusion The structure of 2-aminophenol was optimized by the RHF and DFT methods using the basis sets 6-31 G (d) and cc-pVDZ. Using the optimized geometry, the vibrational frequencies, proton NMR and C13 NMR of the title compound have been calculated and have been found to agree well with experimentally reported values. The small differences are due to the fact that experimental values are recorded in the solid state and theoretical calculations belong to the gaseous phase. A plot of the highest occupied molecular orbital (HOMO) and that of the lowest unoccupied molecular orbital (LUMO) is also made. References
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||